Data access policy note: since January 1st, 2025, registration and authentication are mandatory to access all data curated after this date, either via the web interface or via the application programming interface (API authentication help link). Please contact us if you have any questions.
Whole genome assembly submission
Table of contents
- Does your genome fit the quality requirements ?
- How to make an genome submission ?
- Requesting an embargo period
- Quick links
📚 For further details on submission procedures, please visit BIGSdb documentation website.
Does your genome fit the quality requirements ?
Before submitting genomes, please ensure that they meet the quality criteria defined for the bacterial group. The quality criteria for each bacterial group are available below :| 🦠 Organism | Quality Criteria |
|---|---|
| Bordetella | Quality Criteria |
| Corynebacterium diphtheriae complex | Quality Criteria |
| Elizabethkingia | Quality Criteria |
| Escherichia coli | Quality Criteria |
| Klebsiella pneumoniae species complex | Quality Criteria |
| Leptospira | Quality Criteria |
| Listeria monocytogenes | Quality Criteria |
| Staphylococcus epidermidis | Quality Criteria |
How to make an genome submission ?
- Log onto the database you want to submit data to (ex : Klebsiella, Corynebacterium,...).
- Go to the Isolates and genomes database.

- Click on SUBMISSIONS to access the submission panel.

- Select the genomes submission type.
Alternatively, you can directly access database-specific submission pages using the links in the table.
- Download the template by clicking on one of the following buttons.

- Fill in the template with your info. Copy/paste the template's content into the box then click on SUBMIT.
The metadata fields description for each database is available in the quick links table.
Your assembly files should be named the same as the "assembly_filename" field of the metadata template.
For example, if the isolate name is ATCC13883 in the template, the assembly filename should be ATCC13883.fas.
The suffixes .fa, .fasta or .fna are also accepted.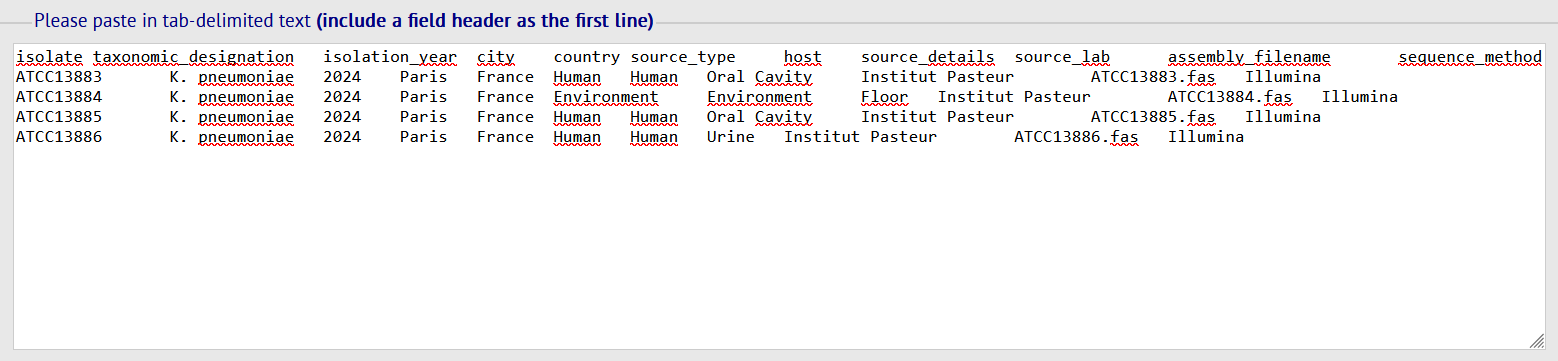
- Upload the assembly files for each isolate using the box provided.
You can drag and drop your files up to 32MB at a time, but one submission can contain up to 500 genomes.
Missing assembly files will be displayed in red. Please make sure that all your genomes have been uploaded correctly before proceeding to the next step.
Assembly file missing Assembly file uploaded 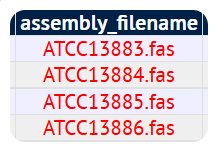

- Make sure everything is correct and that all genomes files have been properly uploaded. Finalize the submission by clicking on the corresponding button.

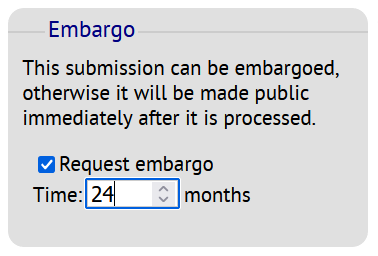
Requesting an embargo period
If you need an embargo period (e.g., prior to publication) you can, when submitting your data, request an embargo by ticking the "Request embargo" box and selecting the desired embargo period.
The request for the initial embargo period cannot exceed 24 months. An embargo period can be extended, more information about it here.
⚠️ Warning : Once the embargo date has passed, the data will be made public immediately. It will no longer be possible to make it private again.
Quick links
| 🦠 Organism | 📝 Metadata description | 📥 Submission page |
|---|---|---|
| Bordetella | Database Fields | Submission page |
| Corynebacterium diphtheriae complex | Database Fields | Submission page |
| Elizabethkingia | Database Fields | Submission page |
| Escherichia coli | Database Fields | Submission page |
| Klebsiella pneumoniae species complex | Database Fields | Submission page |
| Leptospira | Database Fields | Submission page |
| Listeria monocytogenes | Database Fields | Submission page |
| Staphylococcus epidermidis | Database Fields | Submission page |