Data access policy note: since January 1st, 2025, registration and authentication are mandatory to access all data curated after this date, either via the web interface or via the application programming interface (API authentication help link). Please contact us if you have any questions.
Sanger Sequencing Files Submission
🔴 In a nutshell: Sanger file ➡️ Allele submission + Isolate submission
Table of contents
- Why making an isolate submission ?
- How to make an isolate submission ?
- How to make an allele submission ?
- How to make a profile submission ?
- Quick links
📚 For further details on submission procedures, please visit BIGSdb documentation website.
Why making an isolate submission ?
Making an isolate submission is mandatory whether you want to define a new allele or a new MLST profile. This submission is used to collect isolate metadata. If a new allele/MLST profile is defined, the isolate submission also makes the link between the new nomenclature elements and the isolate they originate from.
⚠️ Warning : Please note that if you make an allele/profile submission without an isolate submission, your allele/profile submission will be considered incomplete and will be closed.
How to make an isolate submission ?
- Log onto the database you want to submit data to (ex: Klebsiella, Corynebacterium,...).
- Go to the Isolates and genomes database.

- Click on SUBMISSIONS to access the submission panel.

- Select the isolates submission type.
- Download a template by clicking on one of the following buttons.

- Fill in the template with your info. Copy/paste the template's content into the box then click on SUBMIT.
Metadata fields description are available in the quick links table.
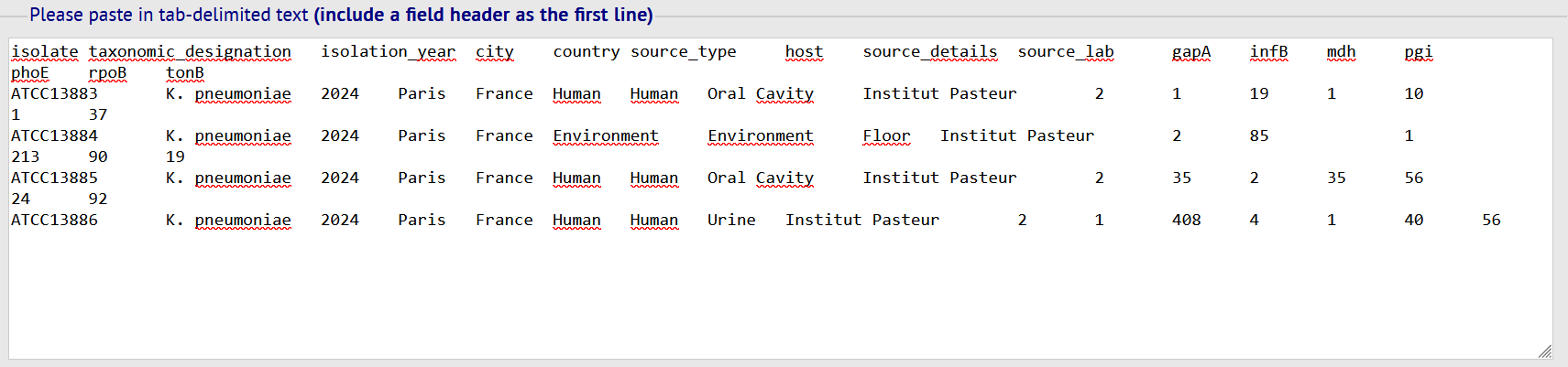
Make sure everything is correct and then finalize the submission by clicking on the corresponding button.

How to make an allele submission ?
Trimming sequences and file naming conventions
You can submit any number of new sequences for a single locus and related trace files.
-
Sequences should be trimmed to the correct start/end sites for the selected locus
-
Trace files should be labeled according to the following model : <1><2><3>_<4>.<5>
- <1> : Strain code (avoiding special characters such as space, dot, slash, etc.)
- <2> : Gene name written exactly as in the database (e.g. gapA)
- <3> : Any additional information you would like
- <4> : Forward Sequencing (F) or Reverse sequencing (R)
- <5> : File extension (.ab1 or .scf)
Example: SB1_gapA_398509385_F.ab1 and SB1_gapA_398509385_R.ab1
⚠️ Warning : Please keep in mind that if any of the previously mentioned steps have not been followed, your submission will be considered incomplete and may be closed.
📌 Note : The discovery of a new allele is always linked to the discovery of a new MLST profile.
Therefore, when submitting a new allele, it is recommended to submit the new MLST profile at the same time. This enables curators to group and process submissions more effectively.
Allele Submission: step-by-step
- Log onto the database you want to submit in (ex : Klebsiella, Corynebacterium,...).
- Go to the Alleles and profiles database.

- Click on SUBMISSIONS to access the submission.

- Select the alleles submission type.

- Choose the locus for which you want to define a new allele, fill in the sequence details fields, provide your new allele sequence then click on SUBMIT.
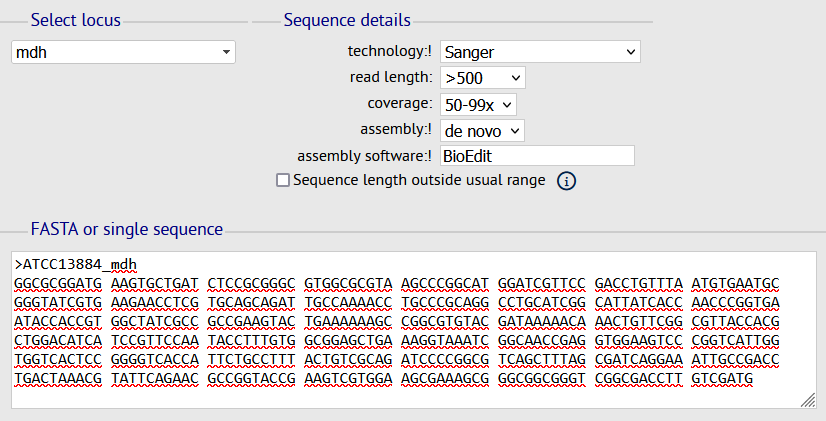
- Upload the Sanger chromatograms (Forward & Reverse) for each new allele submitted using the box provided
- Send your submission by clicking on FINALIZE SUBMISSION

How to make a profile submission ?
Multiple novel profiles can be provided as a single submission.
- Log onto the database you want to submit in (ex : Klebsiella, Corynebacterium,...).
- Go to the Alleles and profiles database.

- Click on SUBMISSIONS to access the submission.

- Select the MLST profile submission type.

- Download a template by clicking on one of the following buttons.

- Fill in the template with your info. Copy/paste the template's content into the box then click on SUBMIT.
The metadata fields description for each database is available in the quick linkstable.
7. You will be redirected to a new page with a summary of the metadata you provided.
Make sure everything is correct and then finalize the submission by clicking on the corresponding button.

Quick links
| 🦠 Organism | Isolates | Alleles | MLST |
|---|---|---|---|
| Bordetella |
Metadata description
Submission page |
Submission page | Submission page |
| Corynebacterium diphtheriae complex |
Metadata description
Submission page |
Submission page | Submission page |
| Elizabethkingia |
Metadata description
Submission page |
Submission page | Not Applicable |
| Escherichia coli |
Metadata description
Submission page |
Submission page | Submission page |
| Klebsiella pneumoniae species complex |
Metadata description
Submission page |
Submission page | Submission page |
| Staphylococcus epidermidis |
Metadata description
Submission page |
Submission page | Not Applicable |