Plugins
Categories - Jump to: Export | Analysis
Export
Export plugins - Jump to: FASTA Export | Profile Export | Sequence Export
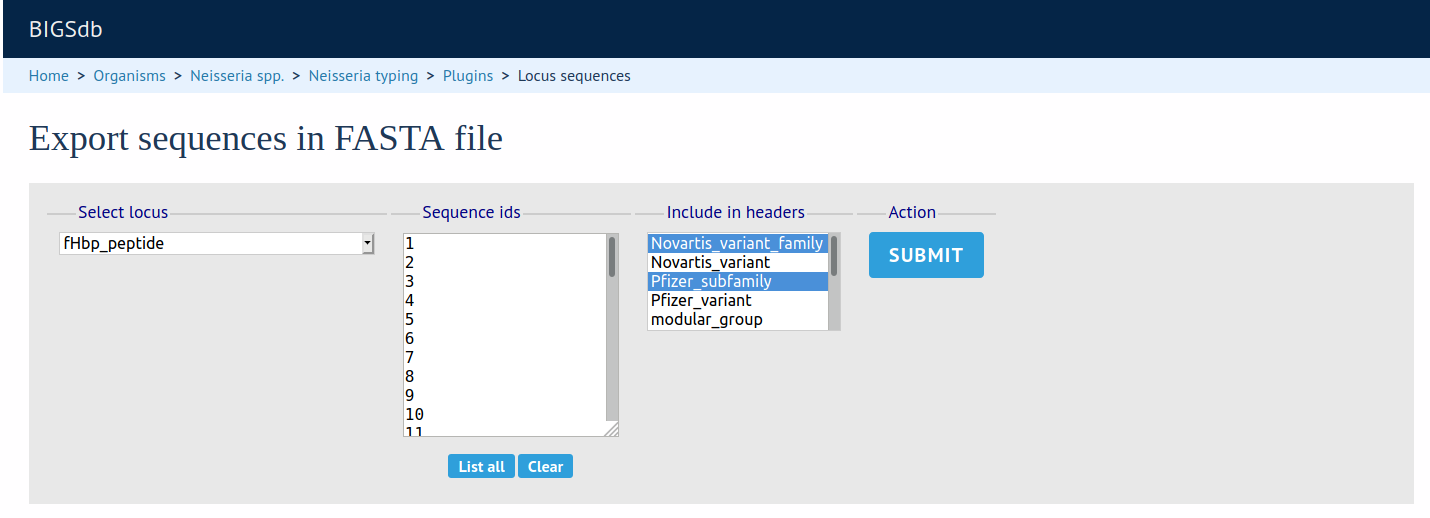
FASTA Export
Summary: Export FASTA file of sequences following an allele attribute query
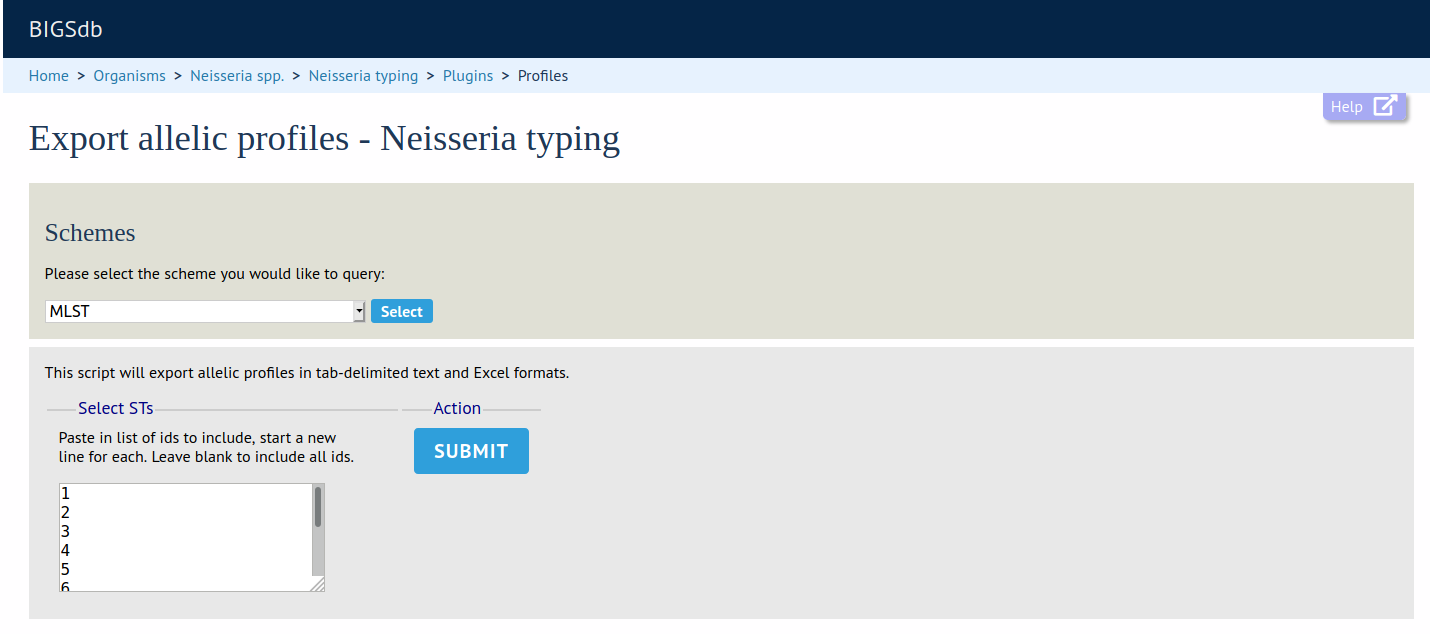
Profile Export
Summary: Export file of allelic profile definitions following a scheme query
Documentation bigsdb.readthedocs.io
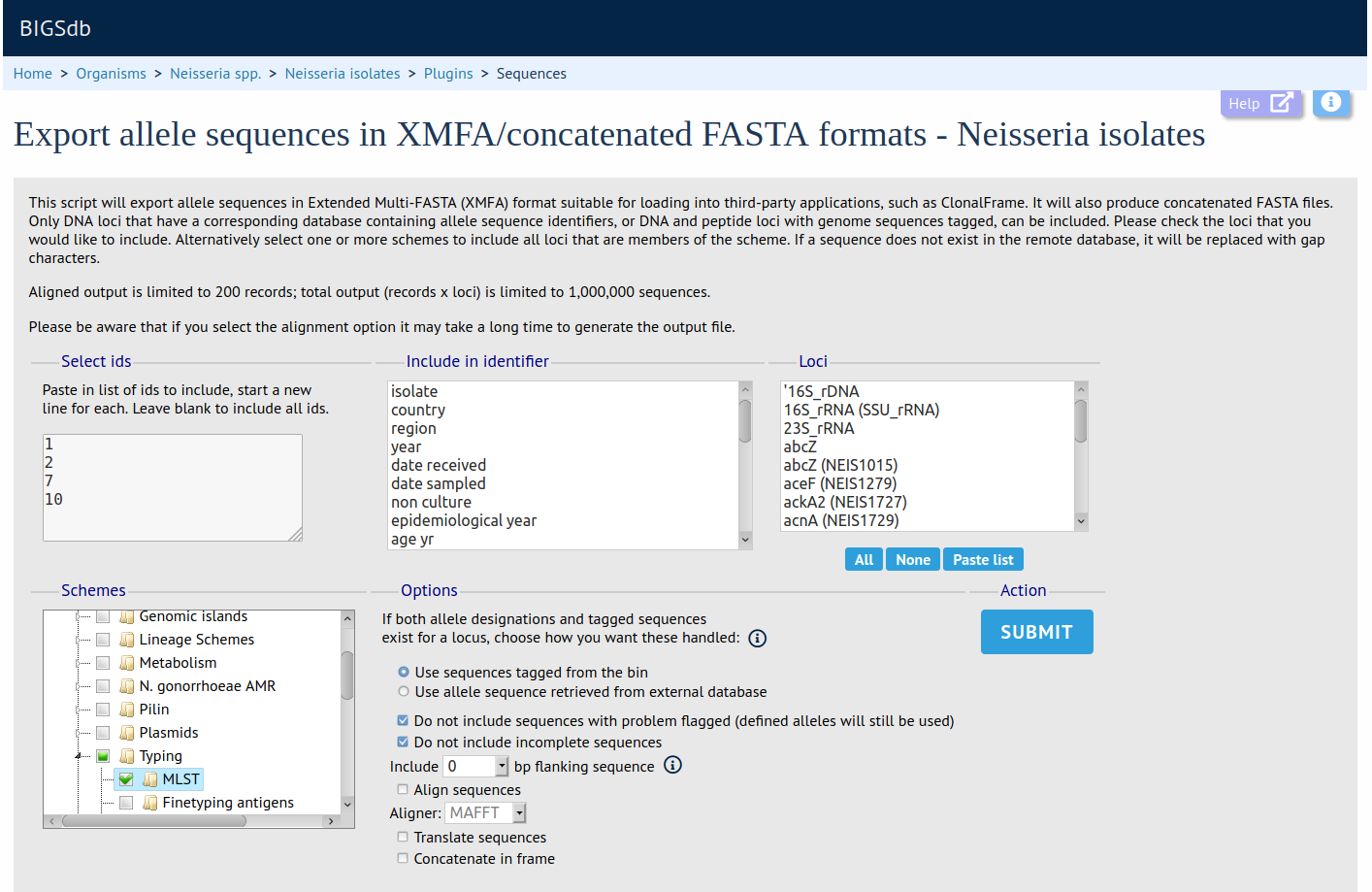
Sequence Export
Summary: Export concatenated allele sequences in XMFA and FASTA formats
This plugin creates concatenated XMFA and FASTA files of selected loci for a particular dataset. These sequences can optionally be aligned, using either MAFFT or MUSCLE, facilitating quick analysis of the outputs in third-party phylogenetic analysis packages.
Documentation bigsdb.readthedocs.io
Analysis
Analysis plugins - Jump to: Sequence Similarity | Sequence Comparison | BURST | Locus Explorer
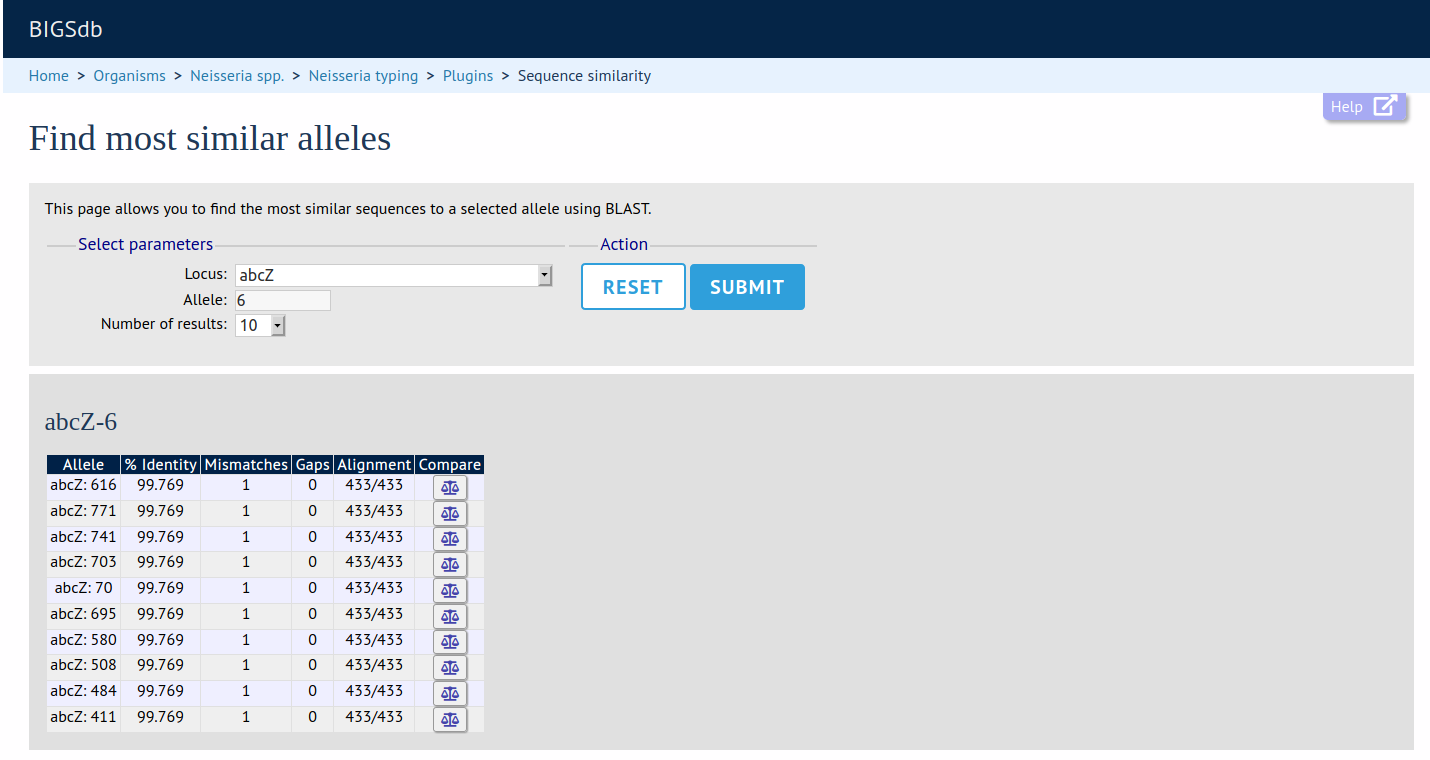
Sequence Similarity
Summary: Find sequences most similar to selected allele
This plugin will return a list of the most similar alleles to a selected allele, along with values for percentage identity, number of mismatches, and number of gaps. Clicking on the link for each returned match will lead to a sequence comparison page, identifying the exact nucleotide/amino acid differences between the query and most similar sequences.
Documentation bigsdb.readthedocs.io
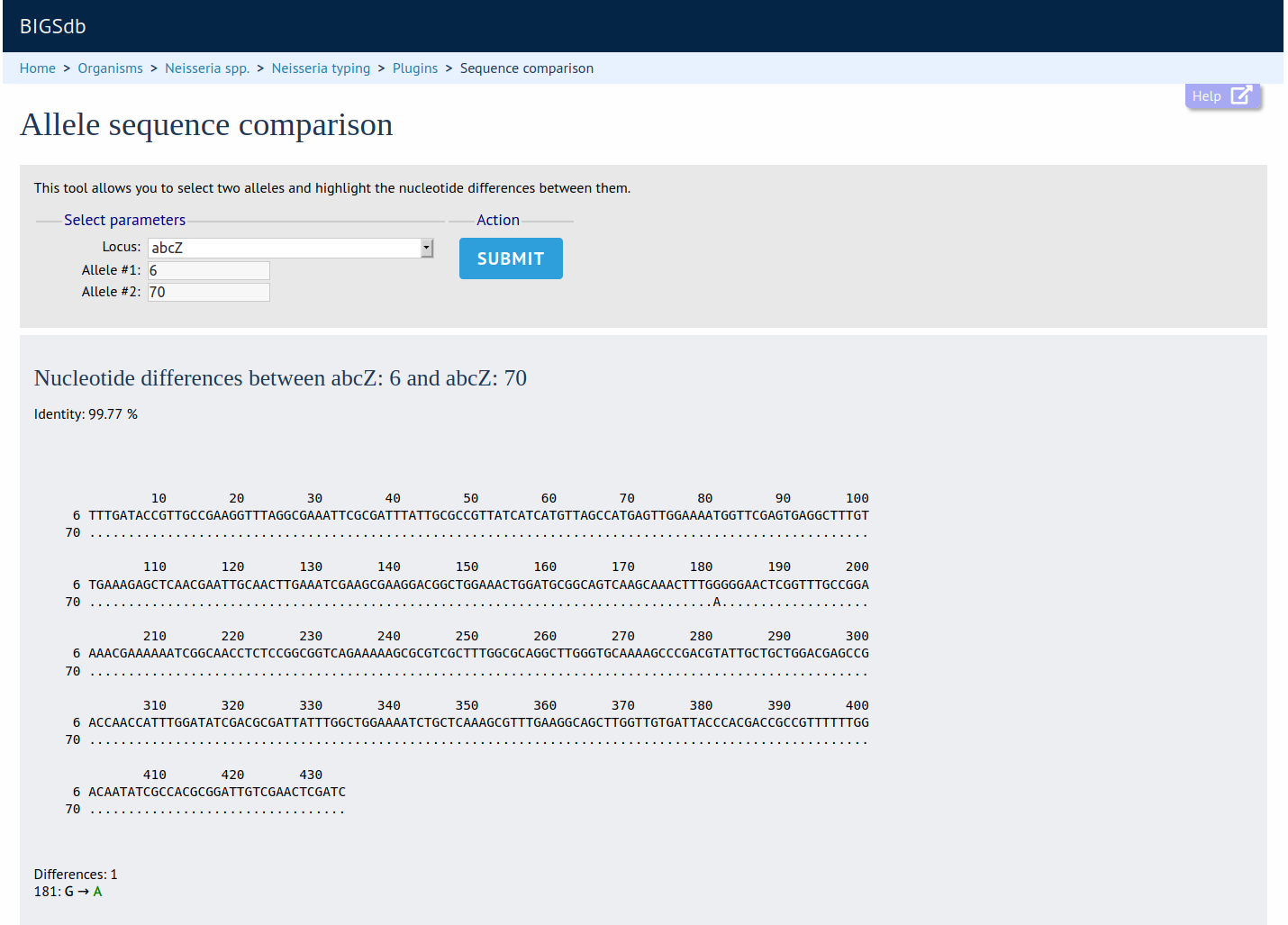
Sequence Comparison
Summary: Display a comparison between two sequences
This shows the nucleotide/amino acid differences between two selected alleles/variants.
Documentation bigsdb.readthedocs.io
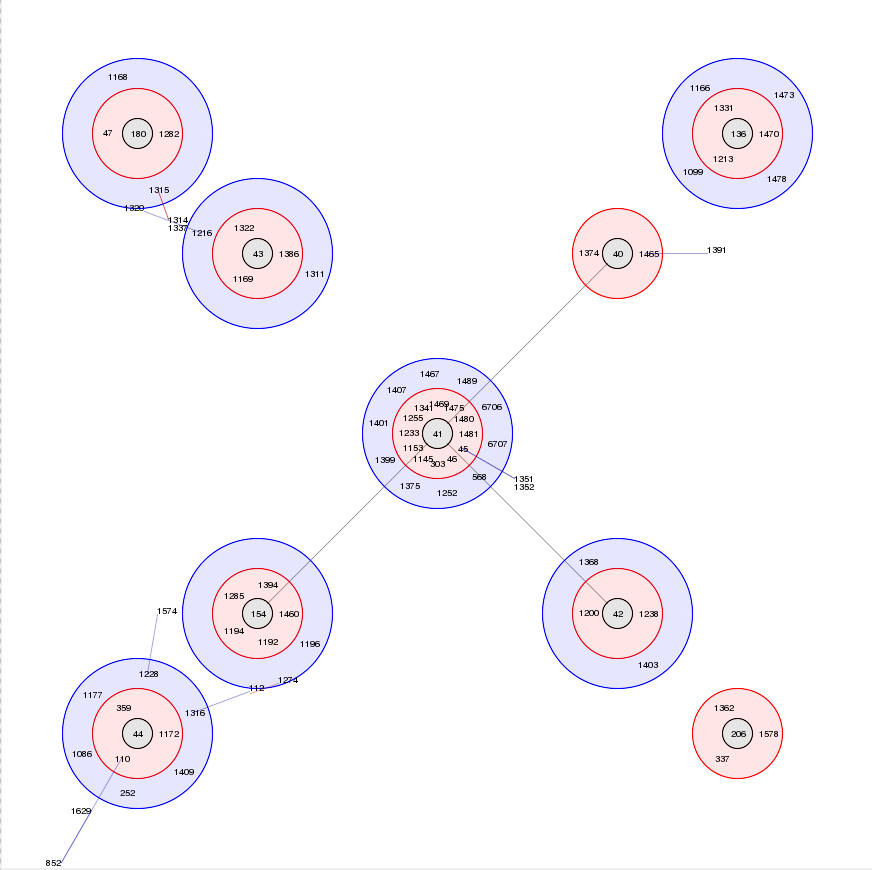
BURST
Summary: Perform BURST cluster analysis on query results query results
BURST is an algorithm used to group MLST-type data based on a count of the number of profiles that match each other at specified numbers of loci. The analysis is available for both sequence definition database and isolate database schemes that have primary key fields set. Analysis is limited to 1000 or fewer records.
Documentation bigsdb.readthedocs.io
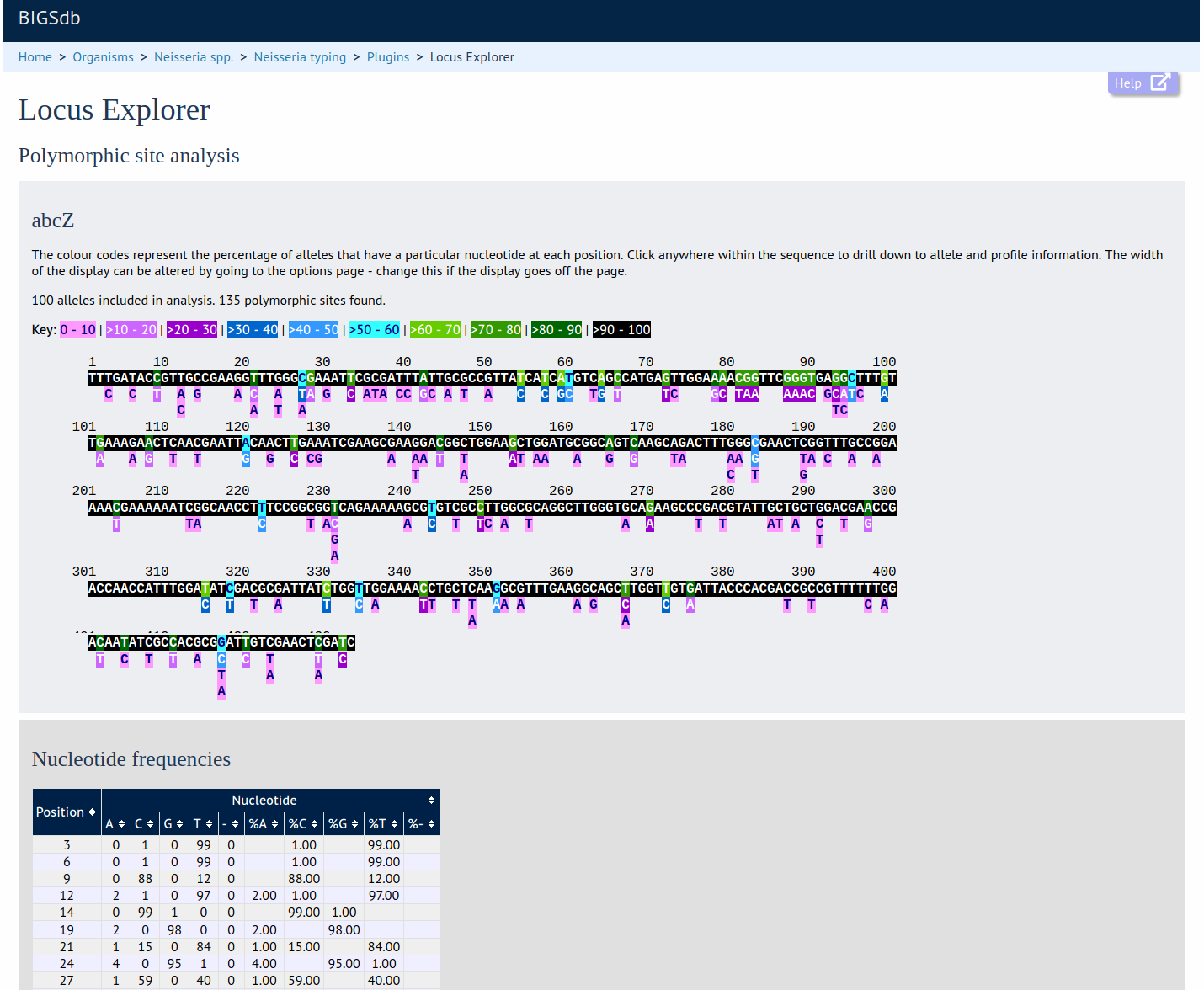
Locus Explorer
Summary: Tool for analysing allele sequences stored for particular locus
This plugin generates a schematic showing the polymorphic sites within a locus, calculate the GC content, codon usage, and generate aligned translated sequences for selected alleles.
Documentation bigsdb.readthedocs.io