Data access policy note: since January 1st, 2025, registration and authentication are mandatory to access all data curated after this date, either via the web interface or via the application programming interface (API authentication help link). Please contact us if you have any questions.
Data Submission Guidelines
📝 Getting an account and registering to databases
If you do not have an account yet, please create one and register it to the database(s) you want to submit to.
Please find a step-by-step guide on the REGISTER page located on the website banner.
⚠️ Warning : Submissions from users with incomplete account information will be rejected.
🔹 NCBI - ENA Submissions
NCBI/ENA and BIGSdb submissions are independent.
When submitting data, if you already have an acccession number you can fill the corresponding metadata field.
Otherwise, you can submit to BIGSdb anytime, and once you have the accession number, please contact us so that we can update your isolate information.
📥 Submit data to BIGSdb-Pasteur
Please find full submission tutorials on BIGSdb documentation website.
We encourage you to submit all the isolates you own so that the database would be as representative of the natural populations as possible. Thus, please do not restrict yourself to submitting only isolates that represent new sequence types : isolates with already known genotypes are also valuable.
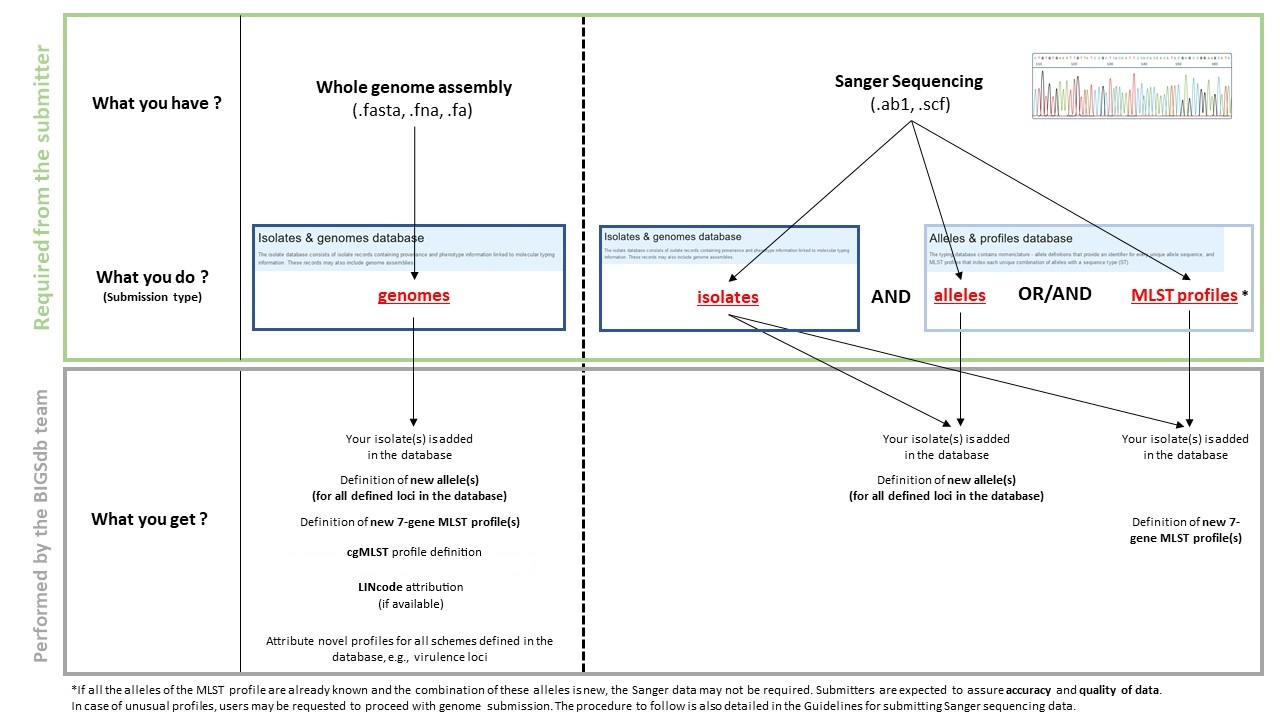
For each pathogen group represented in BIGSdb-Pasteur,two separate databases are available : the Alleles & Profiles database and the Isolates & Genomes database
Depending on your data (whole genomes assemblies or Sanger sequences), you will have to make the required submission(s) in the corresponding database as illustrated on the graph below.
- If you have whole genome assembly files : please follow the whole genome assembly submission guidelines
- If you have Sanger chromatograms : please follow the allele and isolate submission guidelines
- If you want to submit an isolate without genome file : please follow the isolate submission guidelines
- If you want to add a genome to an already existing isolate : please follow the assembly submission guidelines
Once your submission is completed, the curators will automatically receive a notification.
For further information about how BIGSdb-Pasteur handles your data, please read our Policy.
🕜 How long does curation take?
The time required to curate your submission may vary :
- Whole genome assemblies or isolates: results are typically available in less than 48h for small batches or several days for larger batches (hundreds of genomes)
- Sanger data : this may take up to one week to complete the curation process
ℹ️ Data availability
Once the curation is complete, novel alleles and profiles are immediately made available online, as they form the basis of the public nomenclature of genotypes. You will be notified and asked to check if the curated data is correct.
🔒 Data privacy options
Although we encourage the publication of your data as early as possible, all genome submissions (isolates metadata and assemblies) can be kept private. Users can later on make them public and associate them to PUBMED IDs at any given time. Submissions without genome data are immediately made publicly available.
🚩Embargo request
You can request an embargo period to keep your data private for a given amount of time (e.g., prior to the publication of a research paper).
You can do so when submitting your data by ticking the "Request embargo" box and selecting the desired embargo duration. The initial embargo period cannot exceed 24 months (2 years).
⚠️ Warning : Once the embargo period has ended, the data will be made public automatically.
Once public, the data cannot be made private again.
⏳ Extending the Embargo Period
To extend the embargo period, please send a request by email to bigsdb@pasteur.fr at least two months before the current embargo end date.
In your request, please specify the database, the list of isolate BIGSdb IDs and the requested extension duration.
Each extension can be up to a year, with a maximum of three extensions. Hence, your data can remain under embargo for five years at most.
🌍 Publish your data anytime
You can make your isolates public before the embargo date. If you are the owner, you can make a query on the isolates you want to publish and click on the ‘Publish’ button. Once your publication request is confirmed by a curator, your data will become public.
🔄 Data updates
We kindly ask you to contact us in the case of data publication or metadata rectifications. Although data update is possible, please make every effort to make submissions with complete and correct metadata.
📰 Acknowledgments and References
Curation and maintenance of BIGSdb-Pasteur databases is performed by colleagues who dedicate their time to the community.
We'd appreciate if you could acknowledge our efforts by adding the following sentence in the acknowledgments section of your publications, or any other document that might arise from curated data :
We thank the Institut Pasteur teams for the curation and maintenance of BIGSdb-Pasteur databases at https://bigsdb.pasteur.fr/.
In your scientific publications or other documents, please quote the original publications on the development of nomenclature schemes, which can be found on the references page of each organism (see example for Klebsiella references page).
content\databases\klebsiella\subpages\references.md