Data access policy note: since January 1st, 2025, registration and authentication are mandatory to access all data curated after this date, either via the web interface or via the application programming interface (API authentication help link).
To request an API key, follow the steps outlined at: Requesting a BIGSdb-Pasteur API Key
Please contact us if you have any questions.
LIN codes
Quick presentation of LINcode
General presentation
The cgMLST-based Life Identification Numbers (LIN) codes (cgLIN codes, or LIN codes for short) constitute the basis of a novel bacterial strain taxonomy system proposed in 2022. An implementation was initially developed for the Klebsiella pneumoniae species complex (KpSC) by Hennart et al. 2022. This system is a cgMLST adaptation (for the purposes of within-species strain taxonomy) of the universal LIN system proposed by the group of Boris Vinatzer at Virginia Tech University (Marakeby et al. 2014; Tian et al. 2020). The LIN codes are stable by design and their use therefore provides a stable nomenclature (unlike e.g., single linkage classifications, where group fusion can occur).
A LIN code is a multi-position integer-based code attributed to each genome. A cgLIN code consists in a fixed number of ordered bins that determine a partition of the [0%-100%] range of cgMLST profile similarity (Table 1). Each bin is therefore defined by a lower- (inclusive) and an upper- (exclusive) similarity thresholds. The leftmost bins capture the smallest similarities (e.g., deep phylogenetic divisions), whereas the rightmost bins capture the highest similarities (e.g., intended to be useful for epidemiological tracing purposes). The number of positions in a LIN code system should be customized for each taxonomic system (typically, one per bacterial species). cgLIN codes are actually attributed to cgMLST profile sequence types (cgST; derived from genomic sequences), rather than to genomic sequences themselves. Hence, two genomic sequences having the same cgST will have the same LIN code, even when they differ in regions of the genome other than the cgMLST gene sequences. The discriminatory power of LIN codes therefore depends on the nucleotide sequences of the cgMLST scheme upon which it is based.
In the KpSC, a cgLIN code taxonomy was defined using 10 bins Hennart et al. 2022. The four deepest (i.e. first) bins (and their associated ranges and thresholds) were defined considering the phylogenetic structure of the KpSC, based on ~7,000 genomic sequences, and are meant to capture species, subspecies, deep sublineages and clonal groups, respectively (Table 1). The six downstream (i.e. last) bins were defined using thresholds from 10 to 0 cgMLST mismatches (Table 1).
Table 1. Bin identifiers, taxonomic classification and associated ranges/thresholds of the cgLIN coding derived from the KpSC 629-allele cgMLST scheme Hennart et al. 2022
| bin | classification | No. allele mismatches | No. identical alleles | % allele similarity |
|---|---|---|---|---|
| 1 | Species | [629-610[ | [0-19[ | [0.00000-3.02067[ |
| 2 | Subspecies | [610-585[ | [19-44[ | [3.02067-6.99523[ |
| 3 | Sublineage (SL) | [585-190[ | [44-439[ | [6.99523-69.7933[ |
| 4 | Clonal Group (CG) | [190-43[ | [439-586[ | [69.7933-93.1638[ |
| 5 | - | [43-10[ | [586-619[ | [93.1638-98.4102[ |
| 6 | - | [10-7[ | [619-622[ | [98.4102-98.8871[ |
| 7 | - | [7-4[ | [622-625[ | [98.8871-99.3641[ |
| 8 | - | [4-2[ | [625-627[ | [99.3641-99.6820[ |
| 9 | - | [2-1[ | [627-628[ | [99.6820-99.8410[ |
| 10 | - | [1-0[ | [628-629[ | [99.8410-100.000[ |
Each genome (cgSTs, strictly speaking) is encoded following a proximity principle: the LIN code of a new genome j is mainly determined by the LIN code of the most similar genome i already encoded. When two genomes i and j have totally identical cgMLST profiles (i.e., allele similarity sij = 100%), their LIN code identifiers are identical in each of the 10 bins. When sij < 100%, the LIN code of the new genome j is mainly generated from the pivot bin (i.e., the bin whose similarity range includes sij) and the LIN code prefix (determined by the pivot bin) of the closest genome i (see formal process below).
Source database of the LIN codes
For obvious consistency purposes, there can be only one source database for the LIN code definitions. The source of cgLIN codes for KpSC genomes is the BIGSdb-Pasteur sequence definitions database (https://bigsdb.pasteur.fr).
cgLIN code attribution: formal process
First, the allele profile of each genome is calculated based on the 629-loci cgMLST scheme dedicated to the KpSC. As at most 30 missing loci are tolerated, profiles with more than 30 missing alleles are discarded. For each remaining profile, a cgST is defined and next attributed a LIN code in the following way.
The cgLIN code is initialized (i.e., encoded 0 in every bin) with a first allele profile (Table 2). Next, each of the following allele profile j is encoded from the closest profile i (i.e., that maximizes the allele similarity percentage sij) already encoded. After determining the pivot bin p, such that sij ∊ [sp, sp+1 [ (see Table 1), the encoding of the new profile j is performed as follows (see examples in Table 3-6):
(a) same prefix as code i up to the bin p-1 (inclusive);
(b) for the bin p: maximum value observed in this bin among the subset of codes sharing the same prefix incremented by 1;
(c) code 0 in each downstream bin from p+1 (inclusive).
Of note, when sij = 100%, the code of the new profile j is the same as the code of i.
Table 2. Initializing the LIN code with profile 1
| bin = | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| profile 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Table 3. Encoding profile 2 for which the closest is profile 1 (s21 = 618/629 ≈ 98.2512%). As the similarity corresponds to the pivot bin 5 (see Table 1), the prefix is the same as profile 1 up to bin 4 (step a), bin 5 is set to 1 (step b), and each downstream bin from the 6th is set to 0 (step c).
| bin = | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| profile 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| profile 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
Table 4. Encoding profile 3 for which the closest is profile 1 (s31 = 628/629 ≈ 99.8410%). As the pivot bin is 10 (see Table 1), the prefix is the same as profile 1 up to bin 9 (step a), and the 10th bin is set to 1 (step b).
| bin = | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| profile 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| profile 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| profile 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
Table 5. Encoding profile 4 for which the closest is profile 2 (s41 = 628/629 ≈ 99.8410%). As the pivot bin is 10 (see Table 1), the prefix is the same as profile 2 up to bin 9 (step a), and the 10th bin is set to 1 (step b).
| bin = | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| profile 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| profile 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| profile 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| profile 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
Table 6. Encoding profile 5 for which the closest is profile 1 (s51 = 596/626 ≈ 95.2077%). As the pivot bin is 5 (see Table 1), the prefix is the same as profile 1 up to bin 4 (step a), bin 5 is set to 2 (i.e. the largest identifier in bin 5 among the codes sharing the prefix 0_0_0_0 is 1; step b), and each downstream bin from the 6th is set to 0 (step c).
| bin = | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| profile 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| profile 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| profile 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| profile 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| profile 5 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
Notes:
- For two identical cgMLST profiles, the cgLIN code will be the same.
- No cgLIN code is attributed for cgMLST profiles with more than 30 missing alleles (4.77% of the 629 loci), as LIN codes are attributed to cgSTs and as these are not defined in this case.
- The similarity sij between cgST profiles with no more than 30 missing loci is calculated following the formula:
(no. loci with identical alleles in both profiles) / (total no. loci [629] − no. loci with missing allele)
4. For each genome, typically one cgST is attributed based on its cgMLST profile. However, sometimes two (or even more, rarely) cgSTs can match with a given genome (allelic profile), due to the presence of missing data. In these cases, the LIN code of the cgST with the fewest missing data is attributed to the genome.
5. By design, shared cgLIN code prefixes reflect the minimal level of similarity (corresponding to the right border value of the latest bin in the shared prefix) between cgMLST profiles. LIN codes therefore convey a proximity information among genomes (Table 3-6).
6. LIN code attribution is dependent on the order of input into the taxonomy. It was observed (Hennart et al. 2022) that optimal orders (i.e., minimizing the number of identifiers within each bin) are those corresponding to some traversals (e.g. depth-first) of any minimum spanning tree (MStree) inferred from all the pairwise profile distances. When coding a large batch of profiles, it is therefore recommended to order them accordingly (function adjust_prim_order).
A human-readable nomenclature of clonal groups and sublineages: nicknames of LIN code prefixes
The cgLIN codes can be used as a nomenclatural system per se, but it is more convenient to use human-readable and easy to remember nicknames to designate important groups. In the KpSC, all existing prefixes corresponding to the first four levels of the cgLIN codes are attributed aliases (nicknames).
For backward compatibility purposes, these nicknames were chosen based on previously existing nomenclatures:
-
The first bin corresponds to the 5 species of the KpSC (
0_xxxfor K. pneumoniae,1_xxxfor K. variicola,2_xxxfor K. quasipneumoniae,3_xxxfor K. quasivariicola and4_xxxfor K. africana) (Table 7). -
The second bin corresponds to the subspecies (so far, only in K. variicola and in K. quasipneumoniae; the subspecies of K. pneumoniae called rhinoscleromatis and ozaenae are not separate at this deep phylogenetic level – they both belong to K. pneumoniae from a phylogenetic point of view).
Table 7. The correspondence between the two first LIN code levels and Latin ICSP taxonomy
| LIN Prefix | Phylogroup | Phylogroup (sub)species |
|---|---|---|
0_0 | Kp1 | K. pneumoniae |
1_0 | Kp3 | K. variicola subsp.variicola |
1_1 | Kp5 | K. variicola subsp.tropica |
2_0 | Kp2 | K. quasipneumoniae subsp. quasipneumoniae |
2_1 | Kp4 | K. quasipneumoniae subsp. similipneumoniae |
3_0 | Kp6 | K. quasivariicola |
4_0 | Kp7 | K. africana |
- Bins 3 and 4 of the cgLIN codes are the deepest classification levels within species or subspecies, and were arbitrarily called ‘sublineage’ (SL) and ‘clonal group’ (CG) levels, respectively. Since they match well with the classical 7-gene MLST classification, the predominant MLST sequence type within a group at each of these levels (bins) was used as a nickname, where possible (Table 8). A dictionary between MLST identifiers and cgMLST-based classifications was thus created based on an inheritance algorithm (Hennart et al. 2022). For instance, ST258 was the dominant ST among isolates with prefixes
0_0_105and0_0_105_6, and the identifier 258 has therefore been associated to third level prefix0_0_105(which was therefore attributed the nickname SL258) and to fourth level prefix0_0_105_6(nicknamed CG258). See Table 8 for more examples of nicknames of important SL and CG of K. pneumoniae.
Table 8. LIN code prefixes of major SL/CG and corresponding 7-gene MLST classification
| LIN Prefix | SL Nickname | CG Nickname | Majority STs |
|---|---|---|---|
0_0_0_0 | 15 | 15 | 15 |
0_0_429_0 | 23 | 23 | 23 |
0_0_105_6 | 258 | 258 | 258 |
0_0_158_8 | 45 | 45 | 45 |
0_0_197_0 | 147 | 147 | 147 |
0_0_369_0 | 307 | 307 | 307 |
0_0_84_0 | 101 | 101 | 101 |
0_0_1_1 | 14 | 14 | 14 |
Advantages of using the LIN code nomenclature
There are two main advantages of this novel nomenclature:
-
First, the classification into sublineages (SL) and clonal groups (CG) is much more precise than the 7-gene MLST sequence type (ST), as it is based on nearly 100 times more genes. STs do not reflect phylogenetic relationships, as there is no way to know whether two distinct STs are closely related (e.g., ST23 and ST57) or not (e.g., ST258 and ST259). Strains of even the same ST can be distantly related in some cases (e.g., ST23, see Lam et al. 2023). The classification of MLST profiles into ‘clonal complexes’ does not generate relevant groupings, with few exceptions (including the rhinoscleromatis and ozaenae clonal complexes, see Brisse et al. 2009).
-
Second, the human-readable (nicknames) nomenclature of SLs and CGs is stable, as it is based on the LIN code prefixes. It is therefore preferable over the single-linkage classification of cgMLST profiles. This dual taxonomic system with LIN codes and associated prefix nicknames, is related in its outcome (groups are largely overlapping), but distinct in its principle, from the single-linkage classification nomenclature initially proposed in parallel to the LIN codes (Hennart et al. 2022). Single-linkage groupings were quickly abandoned due to the occurrence of fusions as more genomes were introduced.
The implementation of cgLIN codes in PathogenWatch
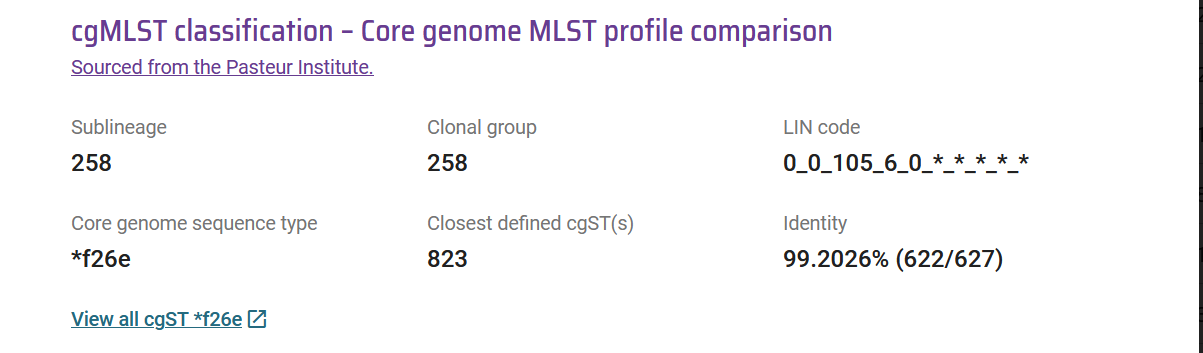
cgLIN codes can be defined in external databases or tools, even though they will often be incomplete as the novel cgMLST alleles, cgST profiles and LIN codes are defined only in the source nomenclature database, BIGSdb-Pasteur (for the KpSC). However, the similarity of a user query genome to already coded ones enable, externally, to infer the genome’s LIN code up to the bin preceding the one in which the similarity value (to a closest encoded genome) falls. If the query genome is closely related to one in the source database, the deduced LIN code will be almost complete (Figure 1). As PathogenWatch uses other ways to provide the phylogroup (species and subspecies), this taxonomic information is not deduced from cgLIN codes in that platform.
PathogenWatch mirrors the defined cgSTs and associated LINcodes from BIGSdb using APIs on a regular basis. Given that PathogenWatch provides provisional alleles, STs and cgSTs, the genome should be submitted inside BIGSdb to obtain definitive identifiers. The provisional information is represented by the asterisk and a code (e.g. cgST *f26e). If a provisional cgST is provided, PathogenWatch also indicates the closest defined cgST. An incomplete cgLIN code is also defined based on the shared prefix obtained from the comparison with the closest reference cgST (Figure 1). This provides information about the relatedness of a query genome compared to the existing taxonomic diversity and can provide sublineage and/or clonal group identification, if LIN code groups of the first four bins are identified (Figure 1).
Figure 1: Example of novel LIN code variant in PathogenWatch
References
Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P (2009) Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One, 4(3):e4982. doi:10.1371/journal.pone.0004982
Hennart M, Guglielmini J, Bridel S, Maiden MCJ, Jolley KA, Criscuolo A, Brisse S (2022) A Dual Barcoding Approach to Bacterial Strain Nomenclature: Genomic Taxonomy of Klebsiella pneumoniae Strains. ** Molecular Biology and Evolution**, 39(7):msac135. doi:10.1093/molbev/msac135
Lam MMC, Holt KE, Wyres KL (2023) Comment on: MDR carbapenemase-producing Klebsiella pneumoniae of the hypervirulence-associated ST23 clone in Poland, 2009–19. Journal of Antimicrobial Chemotherapy, dkad028. doi:10.1093/jac/dkad028.
Marakeby H, Badr E, Torkey H, Song Y, Leman S, Monteil CL, Heath LS, Vinatzer BA (2014) A System to Automatically Classify and Name Any Individual Genome-Sequenced Organism Independently of Current Biological Classification and Nomenclature. PLoS ONE, 9(2):e89142. doi: 10.1371/journal.pone.0089142
Tian L, Huang C, Mazloom R, Heath LS, Vinatzer BA (2020) LINbase: a web server for genome-based identification of prokaryotes as members of crowdsourced taxa. Nucleic Acids Research, 48(W1):W529-W537. doi:10.1093/nar/gkaa190